1. main explanatory variable: cotinine
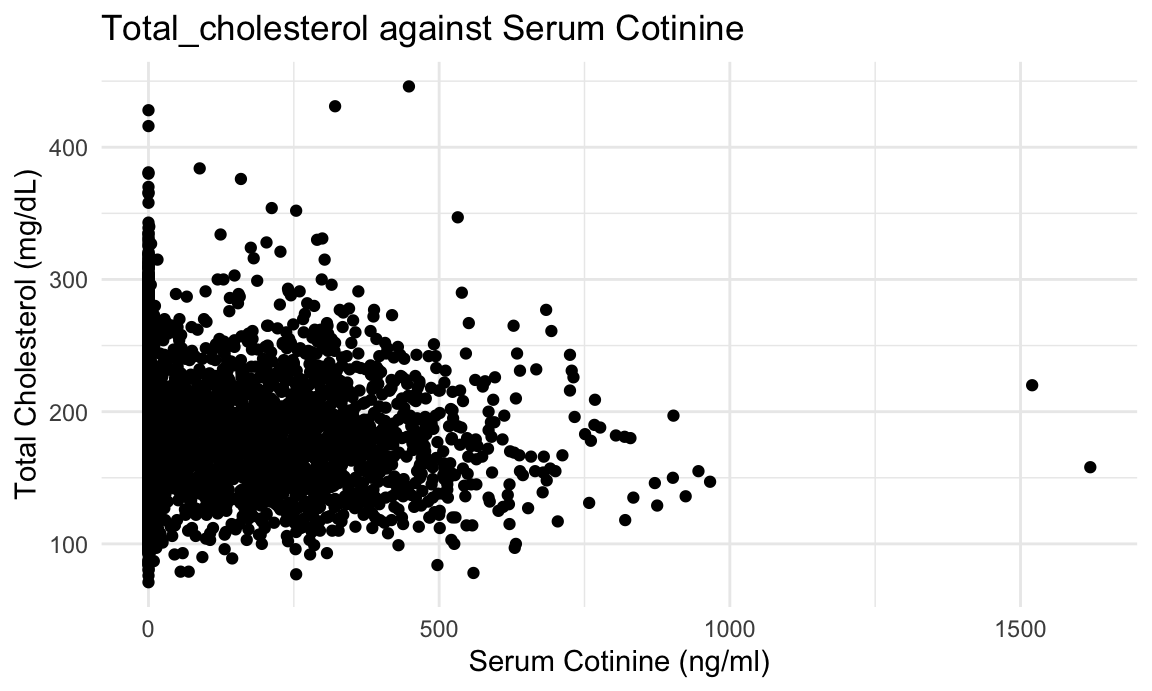
We hypothesize that cotinine, an indicator of exposure of smoking, is positively associated with the log of total cholesterol.
1) Univariable linear regression
fit_1: log(total_cholesterol) = cotinine
fit_1 = lm(log(total_cholesterol) ~ cotinine, data = model_df)
fit_1 |>
broom::tidy() |>
knitr::kable(digits = 3)| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 5.198 | 0.003 | 1944.859 | 0 |
| cotinine | 0.000 | 0.000 | -3.776 | 0 |
model_df |>
modelr::add_residuals(fit_1) |>
ggplot(aes(sample = resid)) +
stat_qq() +
stat_qq_line() +
labs(title = "QQ Plot", x = "Quantile", y = "Residual")
- We can see that cotinine is significantly associated with total cholesterol level. We also check the qq-plot and find that the residuals followed a normal distribution, which indicates a suitability of using linear regression.
- Therefore, we move forward to build multivariable regression.
2) Multivariable linear regression
fit_cot: log(total_cholesterol) = cotinine + age + gender + race + marital_status + education_level_20 + poverty_level
fit_cot = lm(log(total_cholesterol) ~ cotinine + age + gender + race + marital_status + education_level_20 + poverty_level, data = model_df)
fit_cot |>
broom::tidy() |>
knitr::kable(digits = 3)| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 5.221 | 0.015 | 354.487 | 0.000 |
| cotinine | 0.000 | 0.000 | -0.192 | 0.847 |
| age | 0.000 | 0.000 | 1.297 | 0.195 |
| genderMale | -0.045 | 0.006 | -7.911 | 0.000 |
| raceNon-Hispanic Asian | 0.013 | 0.011 | 1.186 | 0.236 |
| raceNon-Hispanic Black | -0.034 | 0.009 | -3.899 | 0.000 |
| raceNon-Hispanic White | -0.012 | 0.008 | -1.463 | 0.144 |
| raceOther Race | 0.009 | 0.014 | 0.607 | 0.544 |
| marital_statusNever married | -0.039 | 0.008 | -4.903 | 0.000 |
| marital_statusWidowed/Divorced/Separated | 0.011 | 0.007 | 1.442 | 0.149 |
| education_level_20College graduate or above | 0.026 | 0.011 | 2.361 | 0.018 |
| education_level_20High school graduate/GED or equivalent | -0.005 | 0.010 | -0.471 | 0.638 |
| education_level_20Less than 9th grade | 0.013 | 0.014 | 0.893 | 0.372 |
| education_level_20Some college or AA degree | 0.010 | 0.010 | 1.006 | 0.314 |
| poverty_levelBelow 130% of Poverty Guidelines | -0.013 | 0.007 | -1.811 | 0.070 |
| poverty_levelBetween 130% and 185% of Poverty Guidelines | -0.002 | 0.008 | -0.214 | 0.831 |
- Based on the estimates of alcohol_use_cat, we can see no association between cotinine and log total cholesterol. This is not consistent with our hypothesis, and the estimate is not significant at 0.05 level of significance.